請在線咨詢或直接聯系17720555249/18986294316
必騰預構載體定制最高100元/個
基本信息
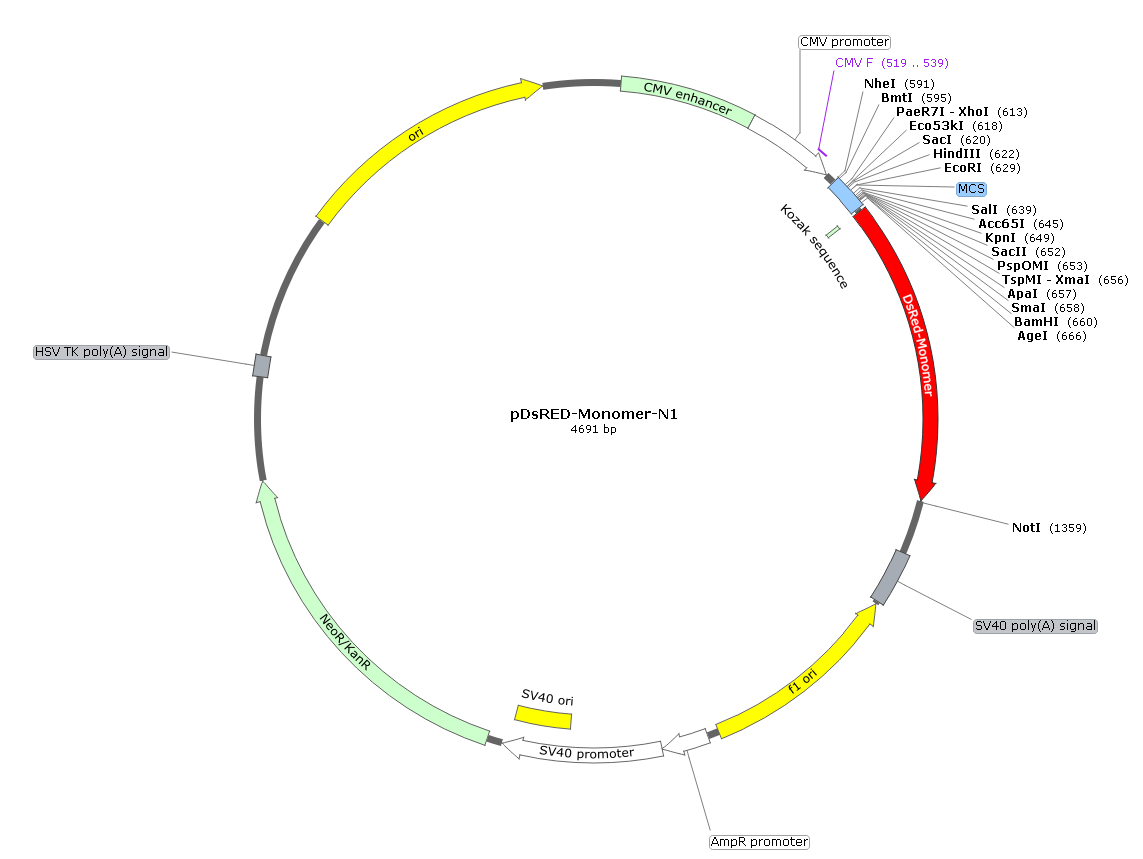
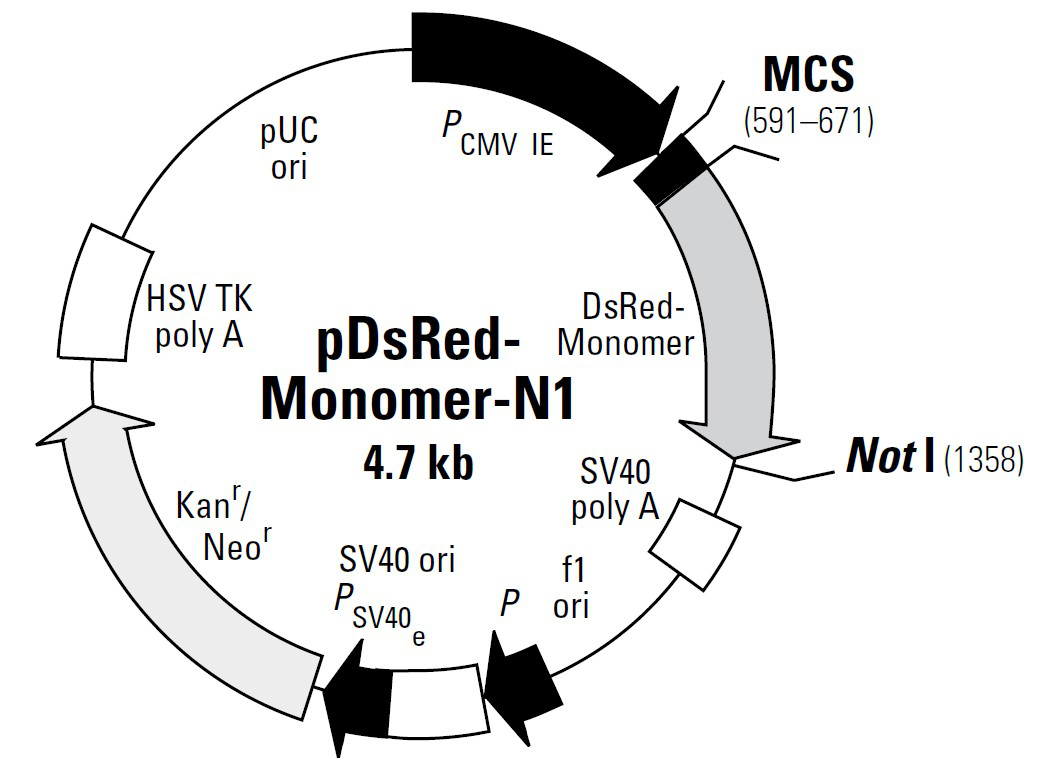
載體類型: 哺乳動物細胞表達載體;紅色熒光報告載體 載體抗性: 卡那霉素 載體標簽: DsRed-Express,紅色熒光蛋白 篩選標記: 新霉素 啟動子: CMV 表達水平: 高拷貝 載體大小: 4691bp 克隆方法: 多克隆位點,限制性內切酶 5' 測序引物: CMV-F: 5’-CGCAAATGGGCGGTAGGCGTG-3’ 3' 測序引物: / 穩定性: 穩表達 或 瞬表達 組成型: 組成型 備注 通過一個轉錄mRNA,能夠同時表達紅色熒光蛋白和一個目的基因。
質粒圖譜和多克隆位點信息
注意:
客戶在我公司選擇構建載體服務項目時,請自行提供母源載體。
如要使用本公司保存的母源載體時,本公司不能保證所有母源載體完全符合客戶預期,也不承擔由此產生的任何責任。
載體描述
pDsRed-Monomer-N1載體是熒光報告載體,表達C端DsRed-Monomer融合蛋白;DsRed-Monomer是DsRed的單體突變體,與DsRed相比,編碼序列經過了人工優化,適用于在哺乳動物細胞中的高水平表達。
pDsRed-Monomer-N1 is a mammalian expression vector that encodes DsRed-Monomer (DsRed.M1), a monomeric mutant derived from the tetrameric Discosoma sp. red fluorescent protein DsRed. DsRed-Monomer contains forty-five amino acid substitutions. When DsRed-Monomer is expressed in mammalian cell cultures, red fluorescent cells can be detected by either fluorescence microscopy or flow cytometry 12–16 hr after transfection (DsRed-Monomer excitation and emission maxima = 557 nm and 592 nm, respectively). The DsRed-Monomer coding sequence is human codon-optimized for high expression in mammalian cells.
DsRed-Monomer is well suited for use as a fusion tag. The multiple cloning site (MCS) in pDsRed-Monomer-N1 is positioned between the immediate early promoter of CMV (PCMV IE) and the DsRed-Monomer coding sequence. Genes cloned into the MCS are expressed as fusions to the N-terminus of DsRed-Monomer if they are in the same reading frame as DsRed- Monomer and there are no intervening stop codons. A Kozak consensus sequence is located immediately upstream of the DsRed-Monomer gene to enhance translational efficiency in eukaryotic systems. SV40 polyadenylation signals downstream of the DsRed-Monomer gene direct proper processing of the 3' end of the DsRed-Monomer mRNA. The vector backbone contains an SV40 origin for replication in mammalian cells expressing the SV40 T antigen, a pUC origin of replication for propagation in E. coli, and an f1 origin for single-stranded DNA production. A neomycin-resistance cassette (Neor) allows stably transfected eukaryotic cells to be selected using G418. This cassette consists of the SV40 early promoter, the neomycin/ kanamycin resistance gene of Tn5, and polyadenylation signals from the Herpes simplex virus thymidine kinase (HSV TK) gene. A bacterial promoter upstream of the cassette confers kanamycin resistance in E. coli.
應用
pDsRed-Monomer-N1 can be used to construct fusions to the N-terminus of DsRed-Monomer. If a fusion construct retains the fluorescent properties of the native DsRed-Monomer protein, its expression can be monitored by flow cytometry and its localization in vivo can be determined by fluorescence microscopy. The target gene must be cloned into pDsRed-Monomer-N1 so that it is in frame with the DsRed-Monomer coding sequence, with no intervening in-frame stop codons. The inserted gene must include an initiating ATG codon. Recombinant pDsRed-Monomer-N1 can be transfected into mammalian cells using any standard transfection method. If required, stable transfectants can be selected using G418. pDsRed-Monomer-N1 can also be used as a cotransfection marker; the unmodified vector will express DsRed-Monomer.
The DsRed1-N Sequencing Primer (Cat. No. 632387) can be used to sequence genes cloned adjacent to the 5' end of the DsRed-Monomer coding region.
For Western blotting, the Living Colors? DsRed Polyclonal Antibody (Cat. No. 632496) can be used to recognize the DsRed-Monomer protein. However, to generate optimal results it may be necessary to use a higher concentration of antibody than recommended on the DsRed Polyclonal Antibody Certificate of Analysis.
載體序列
LOCUS Exported 4691 bp ds-DNA circular SYN 20-MAR-2019
DEFINITION synthetic circular DNA
ACCESSION .
VERSION .
KEYWORDS .
SOURCE synthetic DNA construct
ORGANISM synthetic DNA construct
REFERENCE 1 (bases 1 to 4691)
AUTHORS BT-Lab
TITLE Direct Submission
JOURNAL Exported Wednesday, Mar 20, 2019 from SnapGene Viewer 4.3.4
FEATURES Location/Qualifiers
source 1..4691
/organism="synthetic DNA construct"
/mol_type="other DNA"
enhancer 61..364
/label=CMV enhancer
/note="human cytomegalovirus immediate early enhancer"
promoter 365..568
/label=CMV promoter
/note="human cytomegalovirus (CMV) immediate early
promoter"
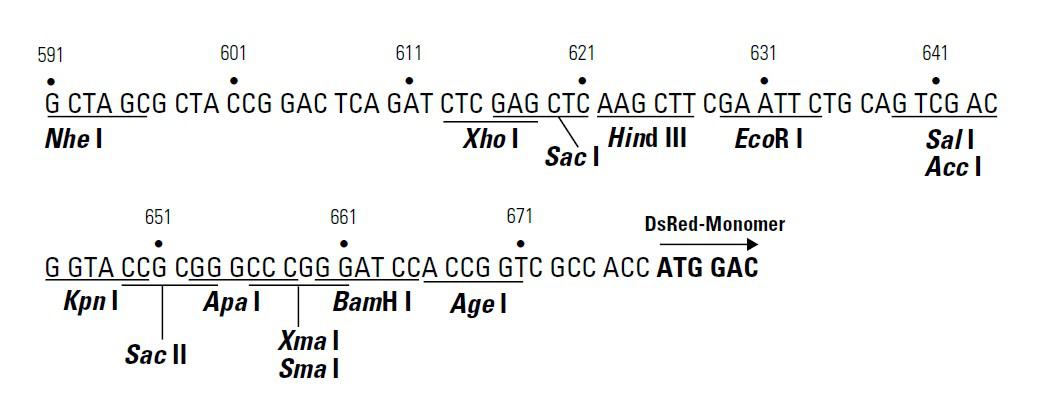
misc_feature 591..671
/label=MCS
/note="multiple cloning site"
regulatory 673..682
/regulatory_class="other"
/note="vertebrate consensus sequence for strong initiation
of translation (Kozak, 1987)"
CDS 679..1356
/codon_start=1
/product="monomeric derivative of DsRed fluorescent protein
(Strongin et al., 2007)"
/label=DsRed-Monomer
/note="mammalian codon-optimized"
/translation="MDNTEDVIKEFMQFKVRMEGSVNGHYFEIEGEGEGKPYEGTQTAK
LQVTKGGPLPFAWDILSPQFQYGSKAYVKHPADIPDYMKLSFPEGFTWERSMNFEDGGV
VEVQQDSSLQDGTFIYKVKFKGVNFPADGPVMQKKTAGWEPSTEKLYPQDGVLKGEISH
ALKLKDGGHYTCDFKTVYKAKKPVQLPGNHYVDSKLDITNHNEDYTVVEQYEHAEARHS
GSQ"
polyA_signal 1479..1600
/label=SV40 poly(A) signal
/note="SV40 polyadenylation signal"
rep_origin complement(1607..2062)
/direction=LEFT
/label=f1 ori
/note="f1 bacteriophage origin of replication; arrow
indicates direction of (+) strand synthesis"
promoter 2089..2193
/gene="bla"
/label=AmpR promoter
promoter 2195..2552
/label=SV40 promoter
/note="SV40 enhancer and early promoter"
rep_origin 2403..2538
/label=SV40 ori
/note="SV40 origin of replication"
CDS 2587..3381
/codon_start=1
/gene="aph(3')-II (or nptII)"
/product="aminoglycoside phosphotransferase from Tn5"
/label=NeoR/KanR
/note="confers resistance to neomycin, kanamycin, and G418
(Geneticin(R))"
/translation="MIEQDGLHAGSPAAWVERLFGYDWAQQTIGCSDAAVFRLSAQGRP
VLFVKTDLSGALNELQDEAARLSWLATTGVPCAAVLDVVTEAGRDWLLLGEVPGQDLLS
SHLAPAEKVSIMADAMRRLHTLDPATCPFDHQAKHRIERARTRMEAGLVDQDDLDEEHQ
GLAPAELFARLKASMPDGEDLVVTHGDACLPNIMVENGRFSGFIDCGRLGVADRYQDIA
LATRDIAEELGGEWADRFLVLYGIAAPDSQRIAFYRLLDEFF"
polyA_signal 3613..3660
/label=HSV TK poly(A) signal
/note="herpes simplex virus thymidine kinase
polyadenylation signal (Cole and Stacy, 1985)"
rep_origin 3989..4577
/direction=RIGHT
/label=ori
/note="high-copy-number ColE1/pMB1/pBR322/pUC origin of
replication"
ORIGIN
1 tagttattaa tagtaatcaa ttacggggtc attagttcat agcccatata tggagttccg
61 cgttacataa cttacggtaa atggcccgcc tggctgaccg cccaacgacc cccgcccatt
121 gacgtcaata atgacgtatg ttcccatagt aacgccaata gggactttcc attgacgtca
181 atgggtggag tatttacggt aaactgccca cttggcagta catcaagtgt atcatatgcc
241 aagtacgccc cctattgacg tcaatgacgg taaatggccc gcctggcatt atgcccagta
301 catgacctta tgggactttc ctacttggca gtacatctac gtattagtca tcgctattac
361 catggtgatg cggttttggc agtacatcaa tgggcgtgga tagcggtttg actcacgggg
421 atttccaagt ctccacccca ttgacgtcaa tgggagtttg ttttggcacc aaaatcaacg
481 ggactttcca aaatgtcgta acaactccgc cccattgacg caaatgggcg gtaggcgtgt
541 acggtgggag gtctatataa gcagagctgg tttagtgaac cgtcagatcc gctagcgcta
601 ccggactcag atctcgagct caagcttcga attctgcagt cgacggtacc gcgggcccgg
661 gatccaccgg tcgccaccat ggacaacacc gaggacgtca tcaaggagtt catgcagttc
721 aaggtgcgca tggagggctc cgtgaacggc cactacttcg agatcgaggg cgagggcgag
781 ggcaagccct acgagggcac ccagaccgcc aagctgcagg tgaccaaggg cggccccctg
841 cccttcgcct gggacatcct gtccccccag ttccagtacg gctccaaggc ctacgtgaag
901 caccccgccg acatccccga ctacatgaag ctgtccttcc ccgagggctt cacctgggag
961 cgctccatga acttcgagga cggcggcgtg gtggaggtgc agcaggactc ctccctgcag
1021 gacggcacct tcatctacaa ggtgaagttc aagggcgtga acttccccgc cgacggcccc
1081 gtaatgcaga agaagactgc cggctgggag ccctccaccg agaagctgta cccccaggac
1141 ggcgtgctga agggcgagat ctcccacgcc ctgaagctga aggacggcgg ccactacacc
1201 tgcgacttca agaccgtgta caaggccaag aagcccgtgc agctgcccgg caaccactac
1261 gtggactcca agctggacat caccaaccac aacgaggact acaccgtggt ggagcagtac
1321 gagcacgccg aggcccgcca ctccggctcc cagtagagcg gccgcgactc tagatcataa
1381 tcagccatac cacatttgta gaggttttac ttgctttaaa aaacctccca cacctccccc
1441 tgaacctgaa acataaaatg aatgcaattg ttgttgttaa cttgtttatt gcagcttata
1501 atggttacaa ataaagcaat agcatcacaa atttcacaaa taaagcattt ttttcactgc
1561 attctagttg tggtttgtcc aaactcatca atgtatctta aggcgtaaat tgtaagcgtt
1621 aatattttgt taaaattcgc gttaaatttt tgttaaatca gctcattttt taaccaatag
1681 gccgaaatcg gcaaaatccc ttataaatca aaagaataga ccgagatagg gttgagtgtt
1741 gttccagttt ggaacaagag tccactatta aagaacgtgg actccaacgt caaagggcga
1801 aaaaccgtct atcagggcga tggcccacta cgtgaaccat caccctaatc aagttttttg
1861 gggtcgaggt gccgtaaagc actaaatcgg aaccctaaag ggagcccccg atttagagct
1921 tgacggggaa agccggcgaa cgtggcgaga aaggaaggga agaaagcgaa aggagcgggc
1981 gctagggcgc tggcaagtgt agcggtcacg ctgcgcgtaa ccaccacacc cgccgcgctt
2041 aatgcgccgc tacagggcgc gtcaggtggc acttttcggg gaaatgtgcg cggaacccct
2101 atttgtttat ttttctaaat acattcaaat atgtatccgc tcatgagaca ataaccctga
2161 taaatgcttc aataatattg aaaaaggaag agtcctgagg cggaaagaac cagctgtgga
2221 atgtgtgtca gttagggtgt ggaaagtccc caggctcccc agcaggcaga agtatgcaaa
2281 gcatgcatct caattagtca gcaaccaggt gtggaaagtc cccaggctcc ccagcaggca
2341 gaagtatgca aagcatgcat ctcaattagt cagcaaccat agtcccgccc ctaactccgc
2401 ccatcccgcc cctaactccg cccagttccg cccattctcc gccccatggc tgactaattt
2461 tttttattta tgcagaggcc gaggccgcct cggcctctga gctattccag aagtagtgag
2521 gaggcttttt tggaggccta ggcttttgca aagatcgatc aagagacagg atgaggatcg
2581 tttcgcatga ttgaacaaga tggattgcac gcaggttctc cggccgcttg ggtggagagg
2641 ctattcggct atgactgggc acaacagaca atcggctgct ctgatgccgc cgtgttccgg
2701 ctgtcagcgc aggggcgccc ggttcttttt gtcaagaccg acctgtccgg tgccctgaat
2761 gaactgcaag acgaggcagc gcggctatcg tggctggcca cgacgggcgt tccttgcgca
2821 gctgtgctcg acgttgtcac tgaagcggga agggactggc tgctattggg cgaagtgccg
2881 gggcaggatc tcctgtcatc tcaccttgct cctgccgaga aagtatccat catggctgat
2941 gcaatgcggc ggctgcatac gcttgatccg gctacctgcc cattcgacca ccaagcgaaa
3001 catcgcatcg agcgagcacg tactcggatg gaagccggtc ttgtcgatca ggatgatctg
3061 gacgaagagc atcaggggct cgcgccagcc gaactgttcg ccaggctcaa ggcgagcatg
3121 cccgacggcg aggatctcgt cgtgacccat ggcgatgcct gcttgccgaa tatcatggtg
3181 gaaaatggcc gcttttctgg attcatcgac tgtggccggc tgggtgtggc ggaccgctat
3241 caggacatag cgttggctac ccgtgatatt gctgaagagc ttggcggcga atgggctgac
3301 cgcttcctcg tgctttacgg tatcgccgct cccgattcgc agcgcatcgc cttctatcgc
3361 cttcttgacg agttcttctg agcgggactc tggggttcga aatgaccgac caagcgacgc
3421 ccaacctgcc atcacgagat ttcgattcca ccgccgcctt ctatgaaagg ttgggcttcg
3481 gaatcgtttt ccgggacgcc ggctggatga tcctccagcg cggggatctc atgctggagt
3541 tcttcgccca ccctaggggg aggctaactg aaacacggaa ggagacaata ccggaaggaa
3601 cccgcgctat gacggcaata aaaagacaga ataaaacgca cggtgttggg tcgtttgttc
3661 ataaacgcgg ggttcggtcc cagggctggc actctgtcga taccccaccg agaccccatt
3721 ggggccaata cgcccgcgtt tcttcctttt ccccacccca ccccccaagt tcgggtgaag
3781 gcccagggct cgcagccaac gtcggggcgg caggccctgc catagcctca ggttactcat
3841 atatacttta gattgattta aaacttcatt tttaatttaa aaggatctag gtgaagatcc
3901 tttttgataa tctcatgacc aaaatccctt aacgtgagtt ttcgttccac tgagcgtcag
3961 accccgtaga aaagatcaaa ggatcttctt gagatccttt ttttctgcgc gtaatctgct
4021 gcttgcaaac aaaaaaacca ccgctaccag cggtggtttg tttgccggat caagagctac
4081 caactctttt tccgaaggta actggcttca gcagagcgca gataccaaat actgtccttc
4141 tagtgtagcc gtagttaggc caccacttca agaactctgt agcaccgcct acatacctcg
4201 ctctgctaat cctgttacca gtggctgctg ccagtggcga taagtcgtgt cttaccgggt
4261 tggactcaag acgatagtta ccggataagg cgcagcggtc gggctgaacg gggggttcgt
4321 gcacacagcc cagcttggag cgaacgacct acaccgaact gagataccta cagcgtgagc
4381 tatgagaaag cgccacgctt cccgaaggga gaaaggcgga caggtatccg gtaagcggca
4441 gggtcggaac aggagagcgc acgagggagc ttccaggggg aaacgcctgg tatctttata
4501 gtcctgtcgg gtttcgccac ctctgacttg agcgtcgatt tttgtgatgc tcgtcagggg
4561 ggcggagcct atggaaaaac gccagcaacg cggccttttt acggttcctg gccttttgct
4621 ggccttttgc tcacatgttc tttcctgcgt tatcccctga ttctgtggat aaccgtatta
4681 ccgccatgca t
//